SUPPLEMENTARY DATA
Evaluation of Short Read Metagenomic Assembly.
By Anveshi Charuvaka (acharuva@gmu.edu) & Huzefa Rangwala(Email: rangwala@cs.gmu.edu)
Dataset Details:
sim36m_dataset_info.csv
(provides the details of the sequences used and the number of reads
taken from each sequence, for simLC, simMC, and simHC dataset)
Replaced Genomes: fames_replace.csv (list of
replacement genomes for the incomplete genomes originally used in Ref
[15] in the paper)
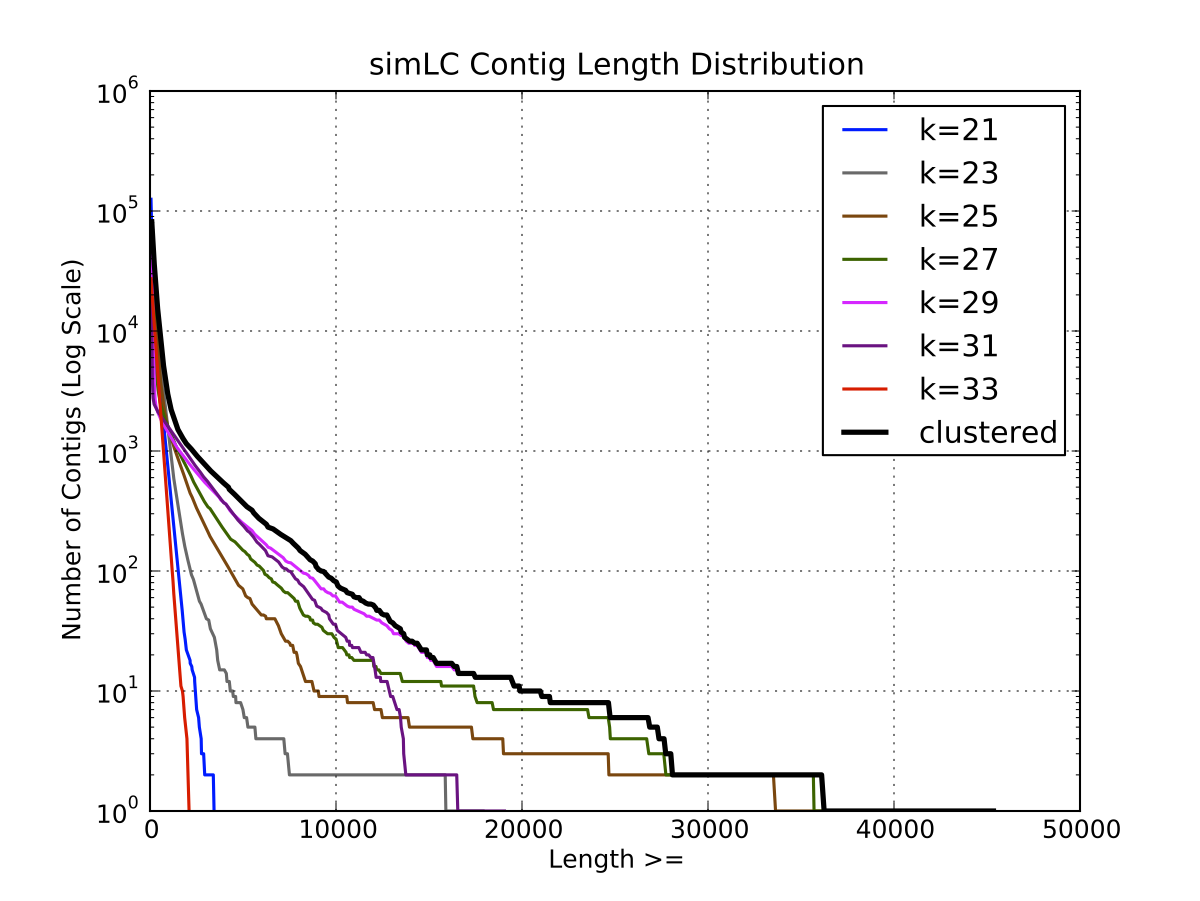
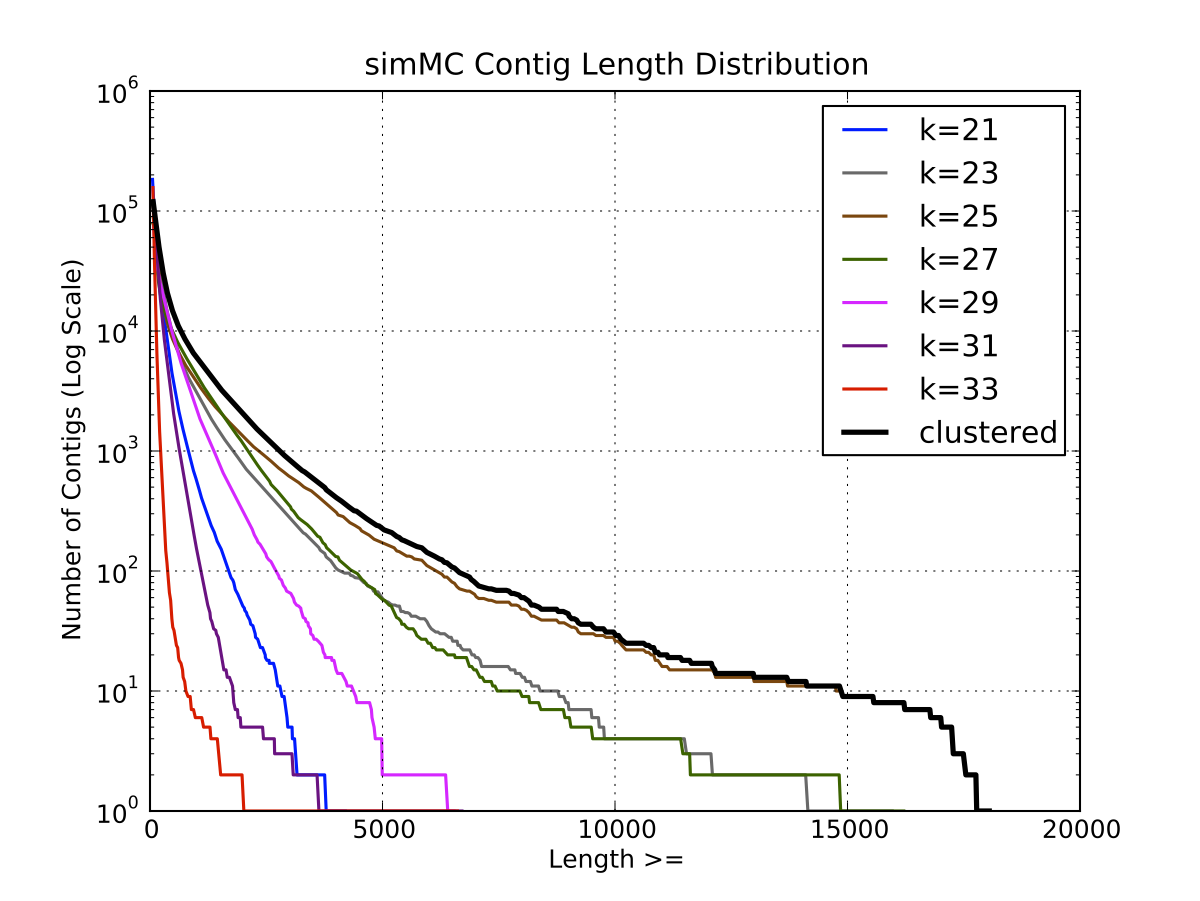
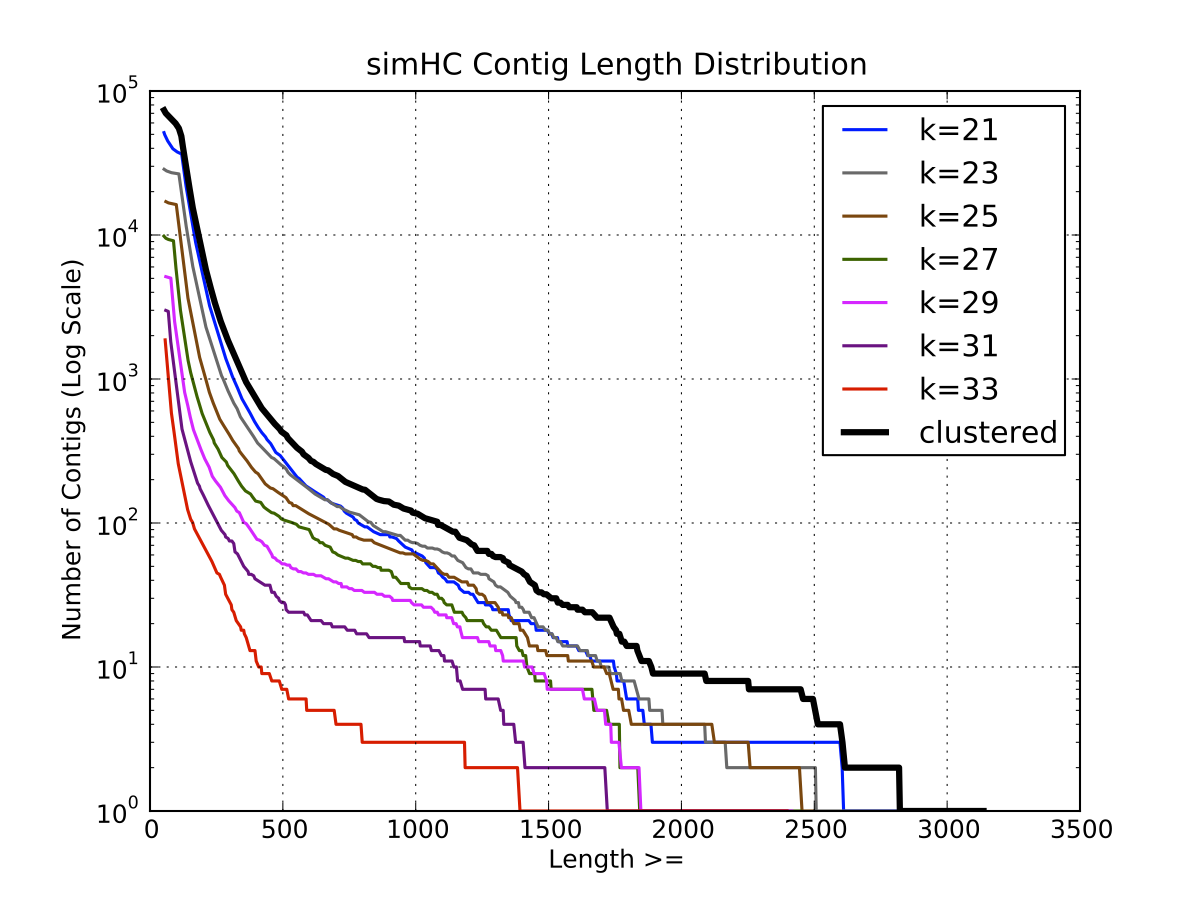
a) Contig Length Distribution
 |
 |
 |
| SimLC Length
Distribution |
SimMC Length
Distribution |
SimHC Length
Distribution |
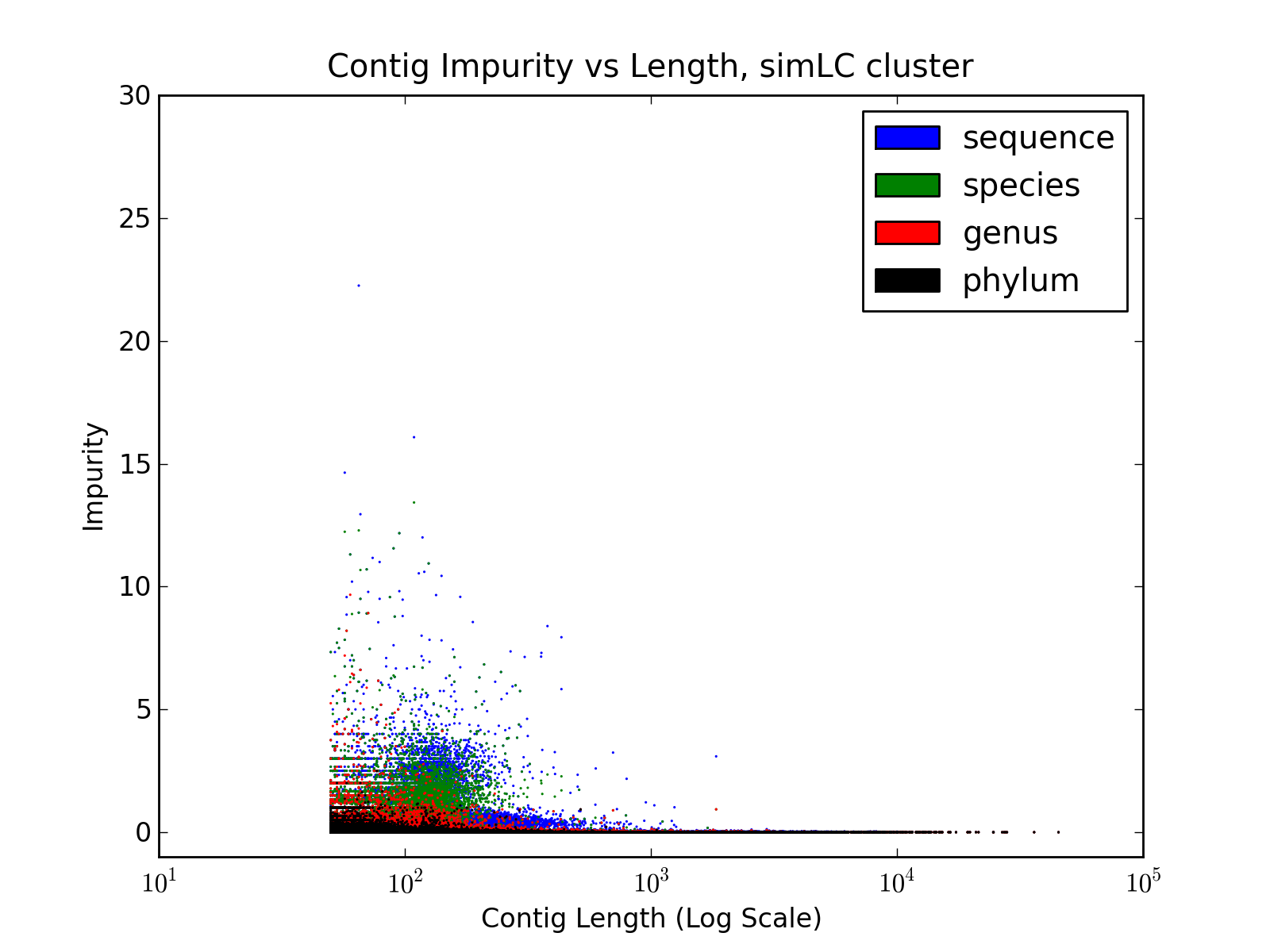
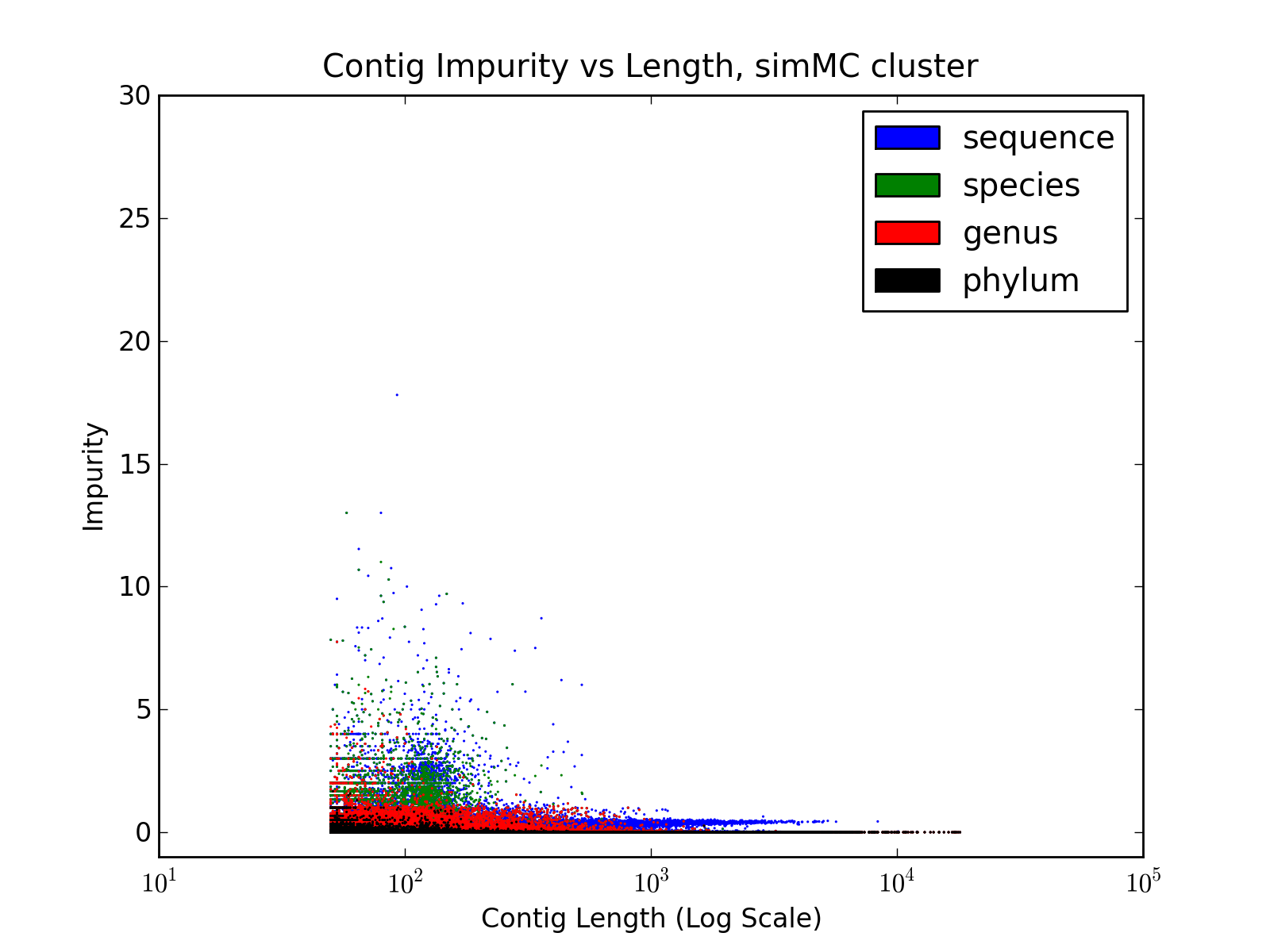
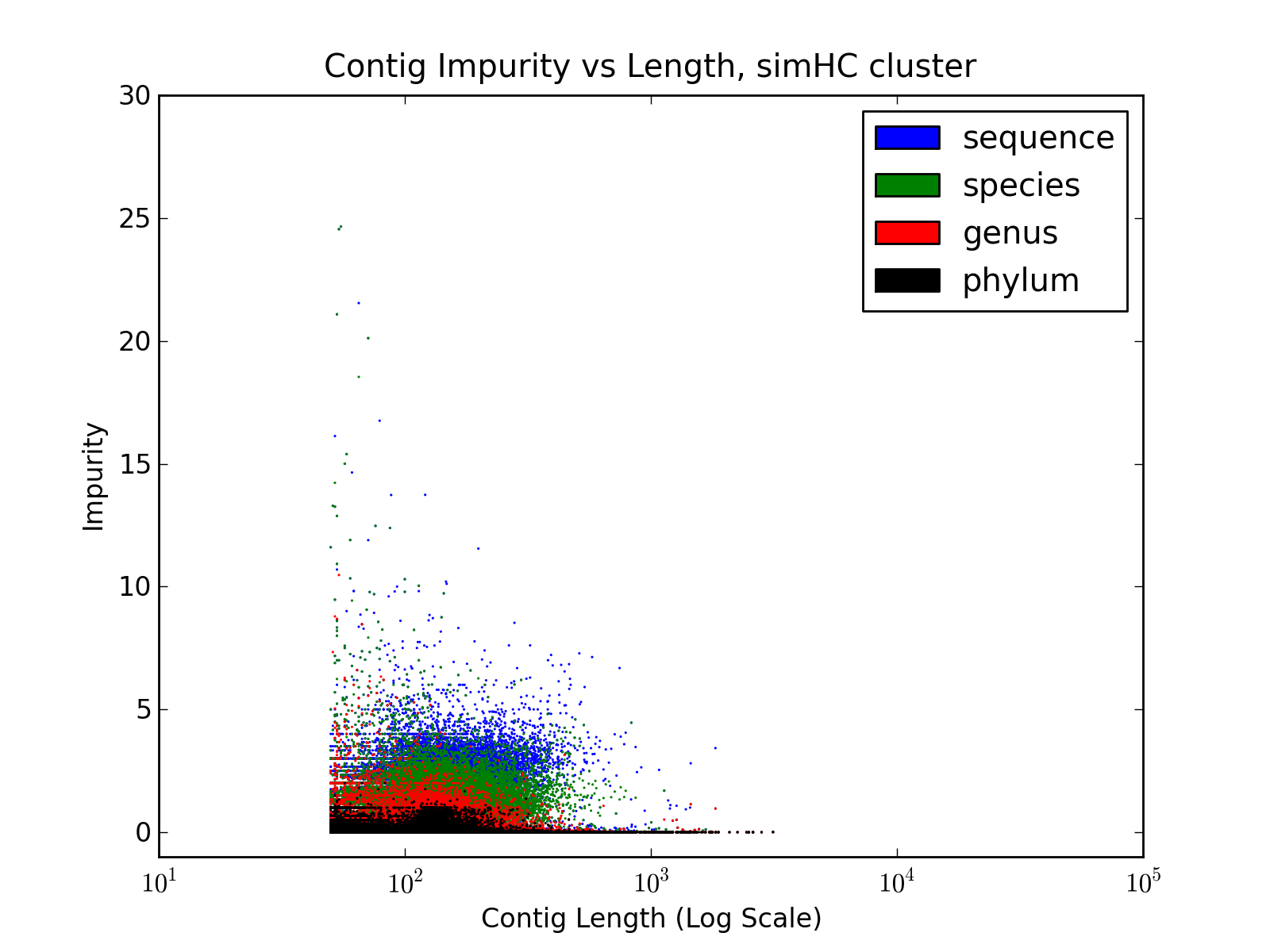
b) Contig Impurity
 |
 |
 |
| SimLC Contig
Impurity |
SimMC Contig Impurity | SimHC Contig Impurity |
Contig Impurity, like Contig Entropy
defined in the paper, represent the degree of chimerism in the contigs.
It is calculated as follows
where b
= max( Ci ) and N = sum( Ci ) ]
- Reads are aligned to the contigs, using BWA, and each read is
mapped to only one contig (to which the read aligns with best accuracy)
- For each contig, the number of reads aligned to it, from different source sequences are counted. Let these counts be denoted by Ci ( where 1 ≤ i ≤ Total Number of Source Sequences )
- Finally, contig impurity is defined as
| Contig Impurity = b /
(N-b) |