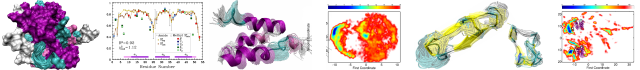Robotics-inspired Search for Modeling Native Structure and Structural Transitions in Proteins
Highlight
of work led by K. Molloyg, B. Olsong
and A. Shehu* at BMC Struct Biol J 2013,
IEEE/ACM Trans Comp Biol and Bioinf (TCBB) 2013, IEEE BIBM-W Comput
Struct Biol Workshop (CSBW) 2012, ACM Conf on Bioinf and Comp Biol
(BCB) 2012, J Bioinf and Comp Biol 2012, IEEE BIBM-W Comp Struct
Biol Workshop (CSBW) 2011, J Bioinf and Comp Biol 2011,
Intl. Conference on Bio-inspired Models of Network, Information, and
Computing Systems (Bionetics) 2010, Intl J Robot Res 2010, Robot Sci
and Sys Conf (RSS) 2009.
The protein conformational space is vast, and the underlying energy surface is rich in local minima. These two characteristics of proteins make it difficult for probabilistic search algorithms to have high exploration (sampling) capability. Specfically, it remains difficult to ensure that computed conformations are geometrically-distinct and not representative of few regions of the conformational space. The difficulty lies in the inability to find a few conformational (reaction) coordinates on which to define distance measures. Popular measures like least Root-Mean-Squared-Deviation (lRMSD) and radius of gyration (Rg), employed in our previous work on modeling native conformations of peptides and proteins, mask away differences and distort locations of regions deemed worthy of further exploration.
This new line of work pursues robotics-inspired search algorithms that capture the connectivity of the conformational space through global graph-like search structures. Lower-dimensional embeddings of the conformational space and underlying energy surface are employed to organize the search space and remember where the search algorithm has been. The centralized search structure allows efficiently computing global statistics and adaptively steering the search to low-energy under-populated regions in the search space, thus reapportioning limited computational resources where needed. Various realizations of this framework have been pursued to model native structures of proteins in the context of decoy sampling and also compute conformational pathways capturing motions of a protein system between two very different functional states.
On this Project:
-
Brian Olson
Kevin Molloy
Amarda Shehu